Plant metabolism is differentiated into primary and secondary metabolism. Secondary metabolites (also known as natural products, phytochemicals or specialized metabolites) underpin important plant traits (e.g. resistance, abiotic stress tolerance, nutritional quality and flavour). The great diversity of secondary metabolites in plants is synthesized from a limited number of building blocks/skeletons that are ubiquitous in the majority of plants but differ in a species-specific manner in the enzymatic permutation and decoration of these basic structures. Thus, despite their common origin a multitude of diverse structures is formed that have divergent biological activities and have traditionally provided a matchless starting point for drug and agrochemical discovery.
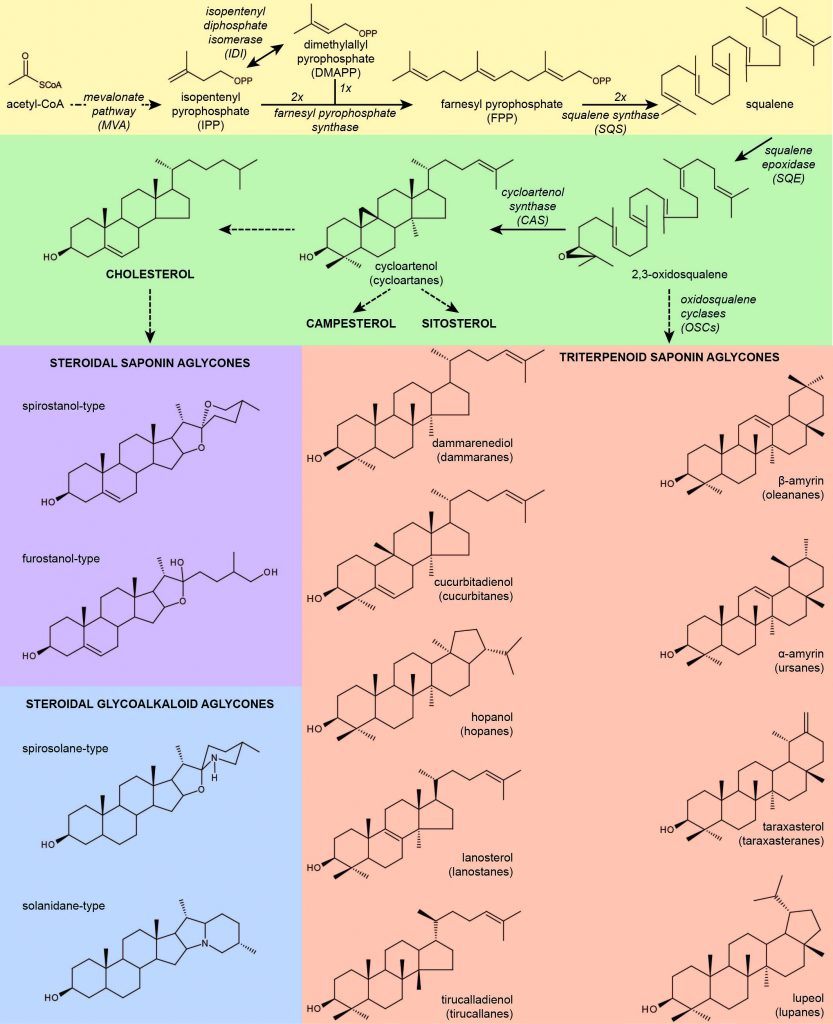
Plant secondary metabolism, focusing on triterpene metabolism, has been a core activity for the last 15 years in the lab. Triterpenes are one of the largest classes of plant natural product and have important functions, both in plants (by providing protection against pests and diseases) and for human use (as drugs, adjuvants, anti-microbials, anticancer agents, surfactants, preservatives etc). Triterpenes are low-molecular weight metabolites that are synthesized from mevalonate via a 30 carbon intermediate, 2,3-oxidosqualene. The cyclization of 2,3-oxido-squalene to sterols and triterpene by enzymes known as oxidosqualenecyclases (OSCs) represents a critical branch-point between primary and secondary metabolism. Yet, the functions of these genes are largely unknown. Work in the lab focuses on triterpene biosynthesis and function in legumes.
Having recently participated in a multi-national, academic-industrial research collaboration, funded by EU FP7 (ΤRIFORC project, www.triforc.eu), to provide new biological tools (genes, enzymes, metabolites) as well as to develop sustainable sources of identified bioactive triterpenes, using combinatorial and synthetic biology approaches in plant and fungal expression systems, we now continue this line of work as part of the OMIC-Engine infrastructure. This infrastructure aims to develop Synthetic Biology research and industrial capabilities in Greece. Within this framework, we are conducting both basic research to elucidate triterpene biosynthetic pathways and their in vivo functions in legumes, and employing these pathways for metabolic engineering in plants and heterologous expression platforms for large-scale triterpene production. As such, projects in the lab involve both traditional molecular biology and genetics approaches, and the development of synthetic biology workflows and tools. Tools under development are in the form of validated biological parts (promoters, markers, other functional sequences) and modules (plasmid vectors) or standardized protocols (such as DNA transformation methods in non-model species). These tools can be employed in legume metabolic engineering, transfer of plant biochemical pathways to heterologous non-plant systems, and legume-optimised targeted genome editing projects.
AMY2 gene cluster project
A novel feature of specialized metabolism in plant biology is that the genes for the synthesis of a number of major classes of plant-derived secondary metabolites, including triterpenes, are organised in clusters, reminiscent of the operons and metabolic gene clusters found in microbes.
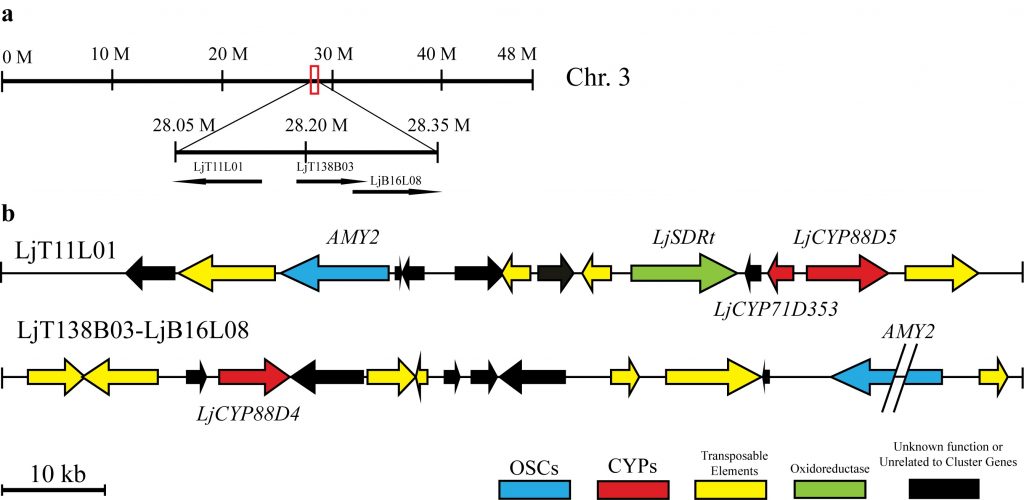
This work focuses on how triterpene biosynthesis is regulated in the model legume, Lotus japonicus, at gene and transcript levels and how it is tailored in response to symbiotic, nitrogen-fixing Rhizobia. Previous work in the lab (Krokida et al., 2013) showed that a multi-functional β-amyrin synthase, AMY2, is clustered with two cytochrome P450 enzymes and an unidentified reductase in the Lotus japonicus genome. AMY2 was strongly co-expressed with both P450 enzymes in response to a range of plant hormone treatments, abiotic stresses, and Rhizobium infection. Current experiments aim to identify the triterpene products of this gene cluster and to elucidate the role of these triterpenes especially in the context of Rhizobium infection. To this end, we have also established a robust root culture system that allows the manipulation of gene expression under controlled conditions.

Additionally, metabolic pathway engineering using the AMY cluster in heterologous expression systems: specially modified E.coli strains, cyanobacteria, and appropriate yeast cells, is ongoing in the lab and in collaboration with other research groups in Greece. The aim of this project is to develop a robust scalable platform, initially for production of a specific triterpene, as a proof or principle, and ultimately to develop a microorganism chassis suitable for expression of oxidosqualene cyclases and downstream tailoring enzymes for production of any triterpene target.
Legume crops
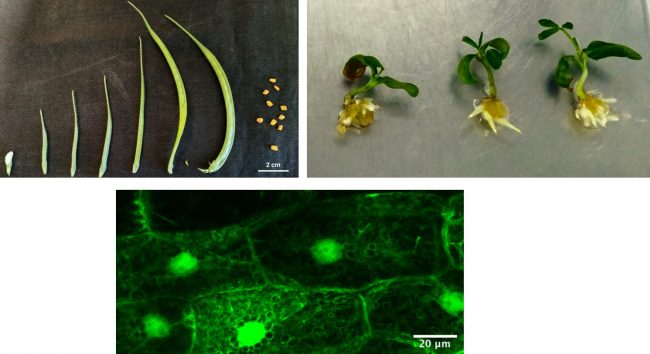
Fenugreek, Trigonella foenum-graecum, is the legume crop under study in our lab. Fenugreek is a forage, food, spice and medicinal plant, producing a high content (0.6-1.7%) and wide array (>95 distinct compounds) of steroidal saponins. As such, it constitutes prime candidate for identification of novel biosynthetic pathways for bioactive or high-value metabolites. However, being a non-model organism, genome sequence information, appropriate genetic transformation tools, and suitable gene promoters and functional DNA sequences are lacking. We have developed a robust and reproducible hairy-root transformation system for Trigonella foenum-graecum and have established culture conditions for fenugreek hairy roots. We are also developing a plasmid vector collection, compatible with modular cloning MoClo/Golden Braid systems. This collection contains existing and newly identified promoters of validated functionality in legumes, and hence in fenugreek. These promoters are also combined with functional DNA sequences for use as selectable markers during fenugreek transformation and for CRISPR/Cas9 genome editing. Apart from developing this toolkit, future work in the lab aims to employ it in metabolic engineering applications in fenugreek and other legumes
Recent Publications
Georgatza D, Gorgogietas VA, Kylindri P, Charalambous MCh, Papadopoulou KK, Hayes JM, Psarra AMG. (2016) The triterpene echinocystic acid and its 3-O-glucoside derivative arerevealed as potent and selective glucocorticoid receptor agonists. The International Journal of Biochemistry &Cell Biology 79 (2016) 277–287
Mosses T, Papadopoulou KK, Osbourn AE (2014) Metabolic and functional diversity of saponins, biosynthetic intermediates and semi-synthetic derivatives. Critical Reviews in Biochemistry and Molecular Biology DOI: 10.3109/10409238.2014.953628
Krokida A, Delis C, Geisler K, Garagkounis C, Tsikou D, Peña-Rodríguez LM, Field B, Osbourn AE, Papadopoulou KK* (2013) A metabolic gene cluster in Lotus japonicus discloses novel enzyme functions and products in triterpene biosynthesis New Phytologist 200: 675-690.
Osbourn AE*, Papadopoulou K, Qi X, Field B, Wegel E. (2012) Finding and analysing plant secondary metabolic gene clusters (Review) Methods in Enzymology 517: 113-138
Delis C, Krokida A, Georgiou S, Peña-Rodríguez LM, Kavroulakis N, Ioannou E, Roussis V, Osbourn AE, Papadopoulou KK (2011) Role of lupeol synthase in Lotus japonicus nodule formation, New Phytologist 189:335-346
Karra A., Konstantinou M., Tzortziou M., Tsialtas I., Kalousi F., Garagounis C., Hayes J., Psarra A.M., Potential Dissociative Glucocorticoid Receptor Activity for Protopanaxadiol and Protopanaxatriol. International journal of molecular sciences, 2019, 20 (1), 94
Garagounis C., Tsikou D., Plitsi K.P., Psarrakou S.I., Avramidou M., Stedel C., Anagnostou M., Georgopoulou M.E., Papadopoulou K.K., Lotus SHAGGY-like kinase 1 is required to suppress nodulation in Lotus japonicus. 2019, The Plant Journal, 98 (2), 228-242
Fatemi, Farzaneh and Abdollahi, Mohammad Reza and Mirzaie-asl, Asghar and Dastan, Dara and Garagounis, Constantine and Papadopoulou, Kalliope; “Identification and expression profiling of rosmarinic acid biosynthetic genes from Satureja khuzistanica under carbon nanotubes and methyl jasmonate elicitation”, Plant Cell, Tissue and Organ Culture (PCTOC), 2019; 136; 561-573.
Funding

Personnel
Dr Constantine Garagounis
Dr Elena Dadami