Functional metagenomics constitute a powerful molecular tool which has facilitated the isolation of novel enzymes of biotechnological interest. Novel pesticide biocatalysts identified via functional metagenomics could find use in environmental protection, human health and agriculture. We have set up a high-throughput pipeline of functional metagenomics, based on an automatic liquid handler, for the isolation of novel pesticide hydrolases from environmental samples with heavy pesticide exposure, i.e. on-farm biodepuration systems like biobeds. Our hypothesis is that in such heavily exposed environments microorganisms are expected to evolve new biodegradation capacities which could be exploited via functional metagenomics. Phenotypic testing of libraries of 20000 clones each resulted in the discovery of novel esterases whose activity against pyrethroids and phenyureas is currently tested.
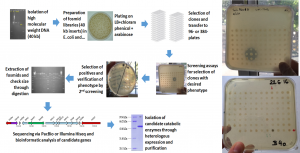
Our functional metagenomics pipeline and phenotypic tests used (tributyrin and X-caprylate) for the isolation of novel pesticide hydrolases from environmental samples
In parallel known catabolic enzymes like PdmAB involved in the demethylation of phenylurea herbicides were isolated through functional metagenomics and their substrate specificity is compared to native PdmAB isolated from soil bacteria. Our hypothesis is that amino acid polymorphisms in the metagenomics-derived PdmAB are associated with a relaxation of the substrate specificity of these enzyme.
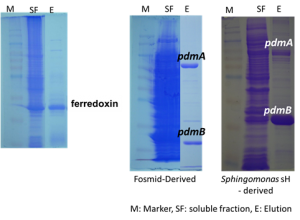
Overexpression and purification of bacteria-derived and metagenomic derived PdmAB and ferredoxin necessary for the optimized function of PdmAB
We utilize synthetic microbial ecology approaches to (i) optimize bioremediation and bioaugmentation efficiency and (b) develop ecosystem relevant synthetic microbial communities for ecotoxicological testing. In this respect we work with wild type and synthetic microbial consortia with optimized biodegradation capacities. We create synthetic microbial consortia composed of bacteria and/or fungi which (i) exhibit nutritional (vitamins and amino acid auxotrophy-based) and pesticide metabolic interdependencies or (ii) exhibit division of labour in a specific function (i.e. nitrification). Regarding wild type consortia we disentangle the mechanisms involved in their coherence and biodegrading function via metagenomics, metatranscriptomics and metaproteomic approaches (see example below).
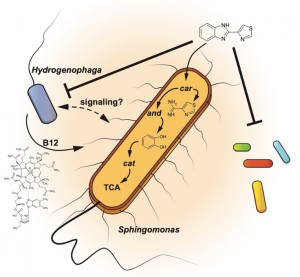
Nutritional interdependencies and the black queen hypothesis driving the function and coherence of a bacterial consortium degrading the fungicide thiabendazole
Funding
 IAPP-MSCA-FP7, Pesticides – Felicity or curse for the soil microbes (LOVE-TO-HATE). Funding: European Commission, 400000 € Duration:2013 – 2016, Coordinator: Dimitrios Karpouzas
IAPP-MSCA-FP7, Pesticides – Felicity or curse for the soil microbes (LOVE-TO-HATE). Funding: European Commission, 400000 € Duration:2013 – 2016, Coordinator: Dimitrios Karpouzas
 Hellenic Research Infrastructure, The research infrastructure of Synthetic Biology in Agro-Nutrition (OMIC-Engine), Funding Body: Hellenic Ministry of Development, Finance and Tourism. Total Funding: 1,600,000 €. Duration 2017-2022
Hellenic Research Infrastructure, The research infrastructure of Synthetic Biology in Agro-Nutrition (OMIC-Engine), Funding Body: Hellenic Ministry of Development, Finance and Tourism. Total Funding: 1,600,000 €. Duration 2017-2022
Publications
- Storck V., Gallego-Blanco S., Vasileiadis S., Begeut J., Rouard N., Hussain S., Baguelin C., Perruchon C., Devers-Lamrani M., Karpouzas D.G., Martin-Laurent F. (2020) Insights into the function and horizontal transfer of isoproturon degrading pdmAB genes in a biobed system. Applied and Environmental Microbiology 86 (14), e00474-20
- Vasileiadis S., Perruchon C., Sheer B., Adrian L., Steinbach N., Trevisan M., Aguera A., Chatzinotas A., Karpouzas D.G., (2021) Nutritional inter-dependencies and a carbazole-dioxygenase are key elements of a bacterial consortium relying on a Sphingomonas for the degradation of the fungicide thiabendazole. BioRxiv doi: https://doi.org/10.1101/2020.03.30.015693
Personnel
Dr Konstantina Rousidou, Postodoctoral fellow
Dr Chiara Perruchon, Postodoctoral fellow